Shengwei Hou
Dr. rer. nat. ( Ph.D.), Bioinformatician & Computational Biologist

Address:
AHF230
3616 Trousdale Pkwy
University of Southern California
Los Angeles, CA 90089
Welcome to my website.
I’m currently doing my postdoc study collaborating with Prof. Jed A. Fuhrman at University of Southern California (USC). Before I moved here in April 2018, I finished my Ph.D. and a short-term postdoc study in Cyanolab supervised by Prof. Wolfgang R. Hess at University of Freiburg in Germany, where I had a curiosity-oriented systematic training and professional development in Computational Biology and Bioinformatics, with a focus on the non-coding elements encoded in three domains of life. Ahead of that, I got a master degree in Genetics at Institute of Hydrobiology, Chinese Academy of Sciences (IHB), supervised by Prof. Xudong Xu on a project of understanding the regulatory roles of genes related to heterocyst development and pattern formation in the multicellular prokaryotic model organism Anabaena sp. PCC 7120. Please check vitae for my detailed curriculum vitae.
I’m currently involved in the “Simons Collaboration on Computational Biogeochemical Modeling of Marine Ecosystems (CBIOMES)” project led by Prof. Michael J. Follows. The CBIOMES project aims to develop mathematical models to quantitatively understand the biogeographical patterns, structures and interactions of diverse marine microbial communities at different space and time scales, and to model the essential traits of marine microbes and their contribution to the global elemental cycles. The Fuhrman Lab is one of the core members of this multidisciplinary collaboration, with a focus on providing marine microbial genetic and genomic information for the modelling purposes. Specifically, the sub-project for me is using genome-resolved metagenomic and metatranscriptomic approaches to understand the distribution and succession of diverse marine microbes and their genetic properties in the context of, but not limited to the “San Pedro Ocean Time-series (SPOT)”.
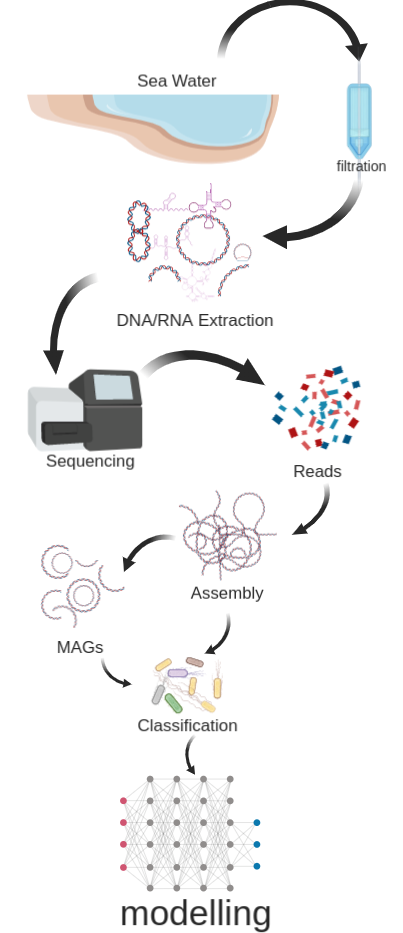
As shown in the following figure (Figure 1), briefly, we
- collect seawater samples from SPOT and the coast of Southern California,
- enrich microbial materials by filtering the seawater through filters,
- extract the whole community DNA and RNA from the filters,
- construct metagenomic and metatranscriptomic libraries for high-throughput sequencing,
- generate assemblies from quality-controlled reads,
- bin assembled contigs into Metagenome Assembled Genomes (MAGs),
- classify microbes into functional categories based on their genetic and transcriptional properties,
- collaborate with modellers to predict their global distribution and contribution to elemental cycles.
news
| Apr 11, 2018 |
I have officially joined the Fuhrman Lab from today |
| Apr 9, 2018 | I got my Ph.D. degree today, a BIG thanks to Wolfgang for supervising me! |